Executive Summary
Siderophores are low molecular weight compounds (500-1500 Da) secreted by microorganisms to chelate and scavenge Fe3+ from the surrounding environment [1]. They are primarily produced by bacteria, fungi, and plants. To date, approximately 500 siderophore structures have been identified [1].
Due to the paucity of Fe3+ in the extracellular environment (a consequence of the low solubility of Fe3+), siderophores often have formation constants (Kf) of 1025 or higher, making them some of the strongest Fe3+ binding agents known [1]. Once a siderophore has bound Fe3+, cells have elaborate mechanisms to detect the siderophore-Fe3+ complex and transport it into the cell. The canonical system for gram-positive (single membrane) bacteria includes a membrane-bound siderophore binding protein (SB) that to the siderophore-iron complex and a siderophore permease, which requires ATP to transport the siderophore across the membrane [2]. Gram-negative (double membrane) bacteria require an additional outer membrane transporter [2].
As a model system of metal binding and transport, siderophores have numerous therapeutic uses within the medical field. For example, native siderophores or mimetic molecules are used for metal chelation, diagnostic imaging, and antibacterial therapies. However, humans also encounter siderophores naturally, such as during an infection.
Contents
1 The necessity and scarcity of soluble iron
2 Structure and Chelation
3 Transport in Gram-Positive Bacteria
3.1 Overview
3.2 Staphylococcus aureus Iron Acquisition
3.3 Staphyloferrin A and SA Transport in S. aureus
3.4 Staphylofferin A and HtsA
4 Siderophores and Medicine
4.1 Common Applications
4.2 Host-Pathogen Interactions
5. Additional Resources
6. References
1. The necessity and scarcity of soluble iron
Iron is required by almost all living organisms, playing crucial roles in cellular processes such as electron transport, transport and storage of O2. It is also a central component of many metabolic enzymes, such as peroxidases, hydrogenases, and nitrogenases [1].
Iron is the fourth most abundant element on Earth. It reacts easily with oxygen leading to the formation of compounds in which iron is in the Fe3+ oxidation state [1]. These iron(III) oxides and hydroxides are frequently responsible for the red or yellow color in many soils and minerals. Iron(III) has incredibly low solubility (Ksp = 10-39), making iron acquisition a challenging for living organisms [1]. Many microorganisms, fungi, and plants have solved the iron acquisition problem via the release siderophores, which are capable of scavenging Fe3+ at sub-femtomolar concentrations from surrounding soil and water [1].
2. Siderophore Structure and Chelation
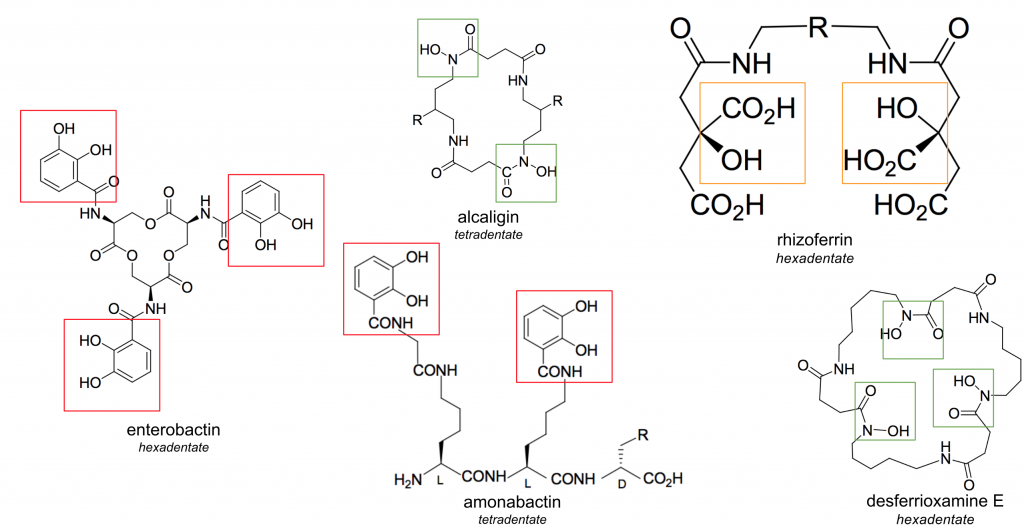
Figure 1. Four siderophore structures of varying denticities and ligand groups (catecholates = red, ⍺-hydroxycarboxylates = orange, hydroxamates = green)
Siderophores are highly varied in structure (Figure 1), yet generally form hexadentate, high-spin, octahedral complexes with Fe3+ [1]. In general, siderophores contain bidentate ligand groups; the most effective siderophores contain three of these groups, allowing them to be hexadentate chelators of iron. Monodentate, bidentate, and tetradentate siderophores do exist, although their stability is generally compromised by the involvement of water molecules in the octahedral complex [1].
Siderophores generally fall into three categories based on the ligands used to chelate iron: catecholates (phenolates), ⍺-hydroxycarboxylates, and hydroxamates (Figure 1, catecholates = red, ⍺-hydroxycarboxylates = orange, hydroxamates = green) [1]. While the most effective ligands coordinate from oxygen atoms, nitrogen and sulfur atoms are occasionally found in ligand groups. These atoms typically reduce the affinity of the siderophore for Fe3+, generally due to distortion of the octahedral field. Regardless of this occasional distortion, siderophores are the strongest known iron chelators. Of the siderophores shown in Figure 1, enterobactin has a Kf of 1049.0, amonabactin has a Kf of 1034.5, desferrioxamine E has a Kf of 1032.5, and rhizoferrin has a Kf of 1025.3 [1]. This high affinity allows siderophores to bind Fe3+ at femtomolar concentrations.
Siderophores universally have a higher affinity for Fe3+ than Fe2+ [1]. This provides a convenient release mechanism via reduction once the siderophore has been transported into the cell. The greater the selectivity for Fe3+ over Fe2+ the more negative the redox potential becomes [1]. While there are many dipositive cations (Zn2+, Cu2+, Ni2+, Mn2+) that are chemically hard to distinguish from Fe2+, Fe3+ is one of the only biologically important tripositive cations, and can be easily selected for over metals like Co3+ and Al3+ [1].
3. Transport in Gram-Positive Bacteria
Overview
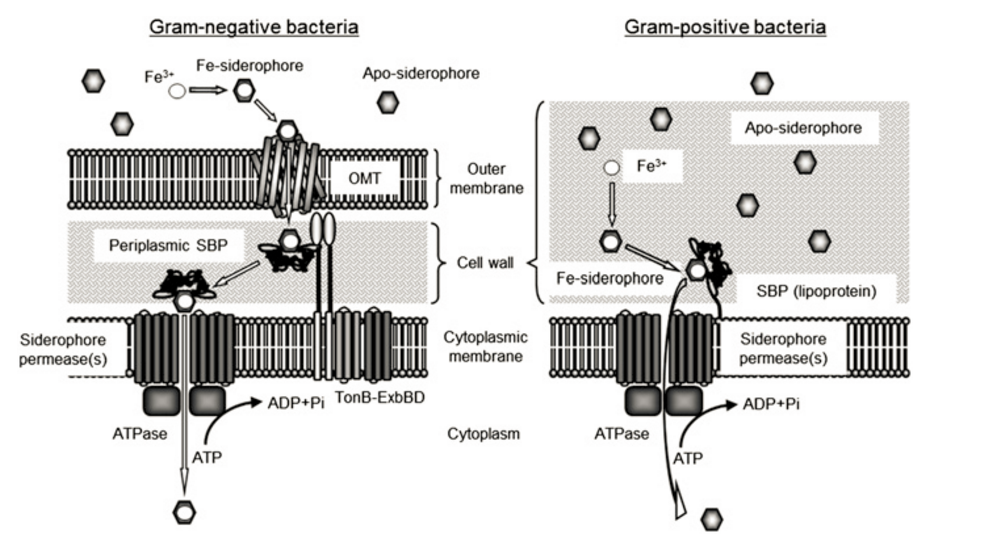
Figure 1. Siderophore Transport in Gram-negative and Gram-positive Bacteria. Image from Fukushima et al. (2013) [3]
Siderophore permeases provide a channel for the Fe-siderophore complex to get into the cell [2]. These belong to a super family of proteins called ABC transporters, named because they contain ATP binding cassettes, which are extra-membranous nucleotide binding domains (NBDs) and provide sites for ATP binding and hydrolysis [2]. Siderophore permeases are usually made up of two homo-dimers, and are not sufficient for Fe-siderophore permeation by themselves, requiring energy from ATP hydrolysis [2].
A third component of the transport system is an ATPase associated with the siderophore permease. The hydrolysis of ATP to ADP and phosphate catalyzed by ATPases provide energy for the siderophore permeases, allowing Fe-siderophore to get into the cell [2].
The main difference between transport in gram-negative and gram-positive bacteria is that since gram-negative bacteria have an additional cellular membrane, an additional outer membrane transporter (OMT) is required. These transporters usually are made up of β-sheets that form a barrel (allowing for the siderophore to get through the membrane), a plug domain that functions as a filter within the barrel, and an extracellular domain that confers specificity for a siderophore [2].
Staphylococcus aureus Iron Acquisition
Staphyloccocus aureus is a round-shaped, gram-positive bacteria that is a major infectious agent in hospitals [4]. About 30% of people carry S. aureus harmlessly, although strains developing antibiotic resistance, such as Methicillin-resistant Staphylococcus aureus (MRSA), have become a significant problem in hospital and community settings [4]. The Center for Diseases Control and Prevention (CDC) estimates that there are 80,000 invasive MRSA infections and over 11,000 deaths related to MRSA every year. S. aureus has diverse mechanisms of Iron acquisition [4].
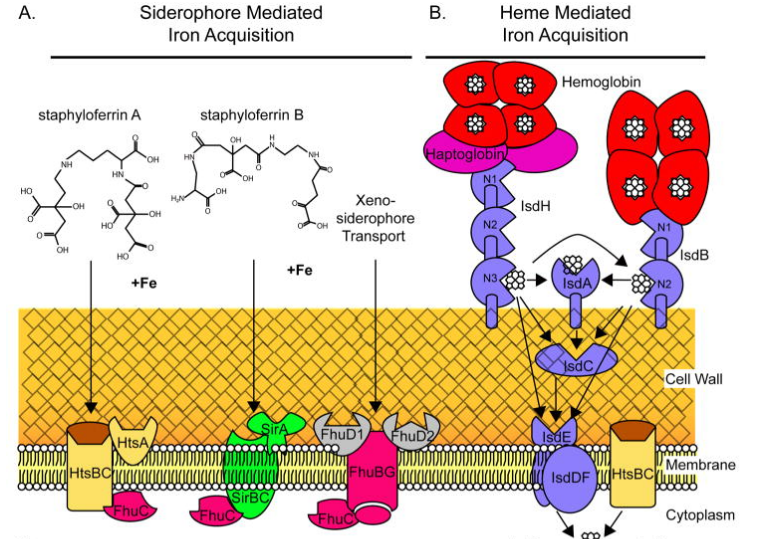
Figure 2. S. aureus Iron Acquisition Systems. Image from Hammer & Skaar (2011) [5]
As heme is the primary Iron source in vertebrates, S. aureus has a heme-mediated iron acquisition system. In fact, in infection settings, heme is the preferred method of iron uptake [5]. Additionally, S. aureus also synthesizes two siderophores of its own, Staphyloferrin A (SA) and Staphyloferrin B (SB), both belonging to the α-hydroxycarboxylate family of siderophores [5]. S. aureus can also take up exogenous sources of siderophores, with most of these siderophores belonging to the hydroxamate group [5].
Staphyloferrin A and SA Transport in S. aureus
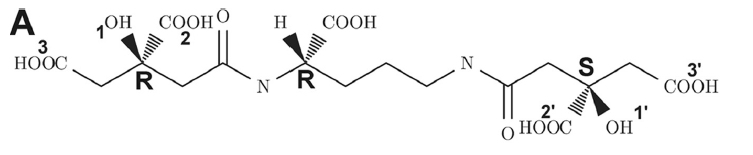
Figure 3. The structure of staphyloferrin A. Figure adapted from Grigg et al. (2010) [6]
Staphyloferrin A transport of iron follows the system typical of gram-positive bacteria. The heme transport system A (HtsA) proteinserves as the siderophore binding protein for staphyloferrin A [6]. Heme transport system BC (HtsBC) serves as the siderophore permease belonging to the ABC transporter family for staphyloferrin A; its associated ATPase is ferric hydroxymate uptake C (fhuC) [6].
Staphyloferrin A and HtsA
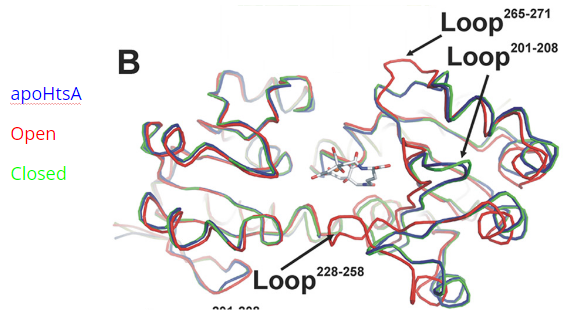
Figure 4. HtsA in the apo-, open, and closed conformations. Figure adapted from Grigg et al. (2010) [6]
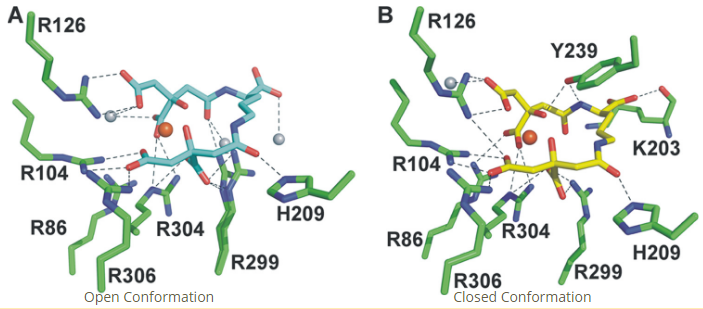
Figure 5. Fe-SA binding in the HtsA binding pocket in the open and closed conformations. Figure adapted from Grigg et al. (2010) [6]
4. Siderophores and Medicine
Therapeutic Applications
Siderophores have numerous applications in the medical field, most of which capitalize upon the their ability to effectively bind and transport metals, for therapeutic use. Siderophore mimetics commonly serve as chelation agents for metal poisoning, iron overload and related diseases [7]. In addition, siderophores are widely used in “Trojan Horse” therapies, whereby a cell’s native transport system is hijacked for the delivery of foreign agents. In these therapies, delivery of non-biologically active metals or antibiotics, typically leads to cell death. Finally, siderophores have been utilized as imaging agents for fungal and bacterial infections. 68Ga-siderophore compounds are gamma radioactive, and can be utilized for positron electron transmission (PET) imaging studies [7].
Host-Pathogen Interactions
Despite their ability to serve as model therapeutics, humans typically encounter siderophores in the context of infection. During infection, host and pathogen compete for a variety of critical resources, including iron. For the pathogen, siderophores serve as the primary iron scavenger. Due to their extremely high iron binding affinity, siderophores have the ability to strip iron from the host’s transport proteins (i.e transferrin, lactoferrin) [7]. In response mammalian hosts secrete two defensive proteins– lactoferrin and siderocalin (Lcn2). Lactoferrin scavenges labile iron, while siderocalin binds siderophore complexes [7]. Figure 6 provides a visual representation of the host-pathogen interplay for iron.
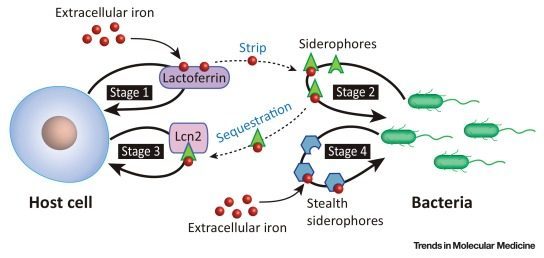
Figure 6. Key players in the host-pathogen “tug-of-war” for iron. Bacteria secrete siderophores to scavenge iron. In response, the host cell defensively sequesters labile and complexes siderophores, via lactoferrin and siderocalin (Lcn2), respectively. Stealth siderophores have features that render them incompatible to siderocalin binding. Image from Wilson et al. (2016).
The siderocalin-enterobactin complex (Scn:[FeIII(Ent)]3-) has been well-characterized and acts as a model system for such a host-pathogen interaction [8]. Siderocalin can bind either apo- or holo-enterobactin with a KD of 0.41nM [8]. The protein’s rigid binding pocket imparts steric specificity, while three positively charged amino acids (R81, K125, K134) intercalate between the enterobactin catechol rings, providing electrostatic stabilization [8]. A hydrogen bond between Y106 and a meta-phenolate as well as cation-pi interactions, provide additional binding interactions [8]. It is hypothesized that upon transport to the endosome the enterobactin backbone is hydrolyzed, yielding 2,3-DHBS [8]. The cleavage is predicted to increase the redox potential of the complexed Fe3+, potentially allowing for host retrieval of Fe2+ with the aid of ferrireductase or an equivalent reducing enzyme [8].
The steric and electrostatic components of the siderocalin binding pocket are suited for catechol-based siderophore complexation. Therefore, ⍺-hydroxycarboxylate or hydroxamate type siderophores typically escape such an immune response. While some pathogens have always produced siderophores including these alternative moieties, several others have adopted modified mechanisms of siderophore synthesis with the intention of siderocalin evasion. An example of such a “stealth siderophore” is anthrax pathogen’s petrobactin. Petrobactin, which is originally synthesized from a catechol-based precursor, exhibits 3,4-phenolate functionality that is sterically incompatible with the siderocalin binding pocket [9].
If you’re interested in reading more about petrobactin and siderocalin evasion, check out this article.
5. See also
**VOTED BEST SOURCE** The video we found:
This video provides a good overview of what siderophores are, their structures, and how they function. https://www.youtube.com/watch?v=l2_3ZMQUKC0
A quizlet on some basic facts of siderophores: https://quizlet.com/72569755/siderophores-and-stuff-flash-cards/
A brief textbook chapter with a quiz: https://www.boundless.com/microbiology/textbooks/boundless-microbiology-textbook/cell-structure-of-bacteria-archaea-and-eukaryotes-4/transport-across-the-cell-membrane-33/siderophores-257-7502/
Siderocalin as the PDB “Molecule of the Month”: https://pdb101.rcsb.org/motm/193
6. References
[1] R.C. Hider and X. Kong. “Chemistry and biology of siderophores.” Natural Product Reports. 2010, 27(5), 637-657.
[2] Faraldo-Gómez, J.D. (2007). “Protein-mediated Siderophore Uptake in Gram-negative Bacteria: A Structural Perspective”, Microbial Siderophores, Volume 12, pp 105-120.
[3] Fukushima T, Allred BE, Sia AK, Nichiporuk R, Andersen UN, Raymond KN (2013) Gram-positive siderophore-shuttle with iron-exchange from Fe-siderophore to apo-siderophore by Bacillus cereus YxeB. Proceedings of the National Academy of Sciences of the United States of America. 110(34):13821-13826. doi:10.1073/pnas.1304235110.
[4] Center for Disease Control and Prevention. Staphylococcus aureus in Healthcare Settings. 2017https://www.cdc.gov/hai/organisms/staph.html
[5] Hammer, N. D., & Skaar, E. P. (2011). Molecular mechanisms of Staphylococcus aureus iron acquisition. Annual Review of Microbiology, 65, 10.1146/annurev–micro–090110–102851. http://doi.org/10.1146/annurev-micro-090110-102851
[6] Grigg JC, et al (2010). “The Staphylococcus aureus Siderophore Receptor HtsA Undergoes Localized Conformational Changes to Enclose Staphyloferrin A in an Arginine-rich Binding Pocket” The Journal of Biological Chemistry, vol 285, no. 15, 11162–11171
[7] Wilson, B. R., Bogdan, A. R., Miyazawa, M., Hashimoto, K., Tsuji, Y.; Siderophores in Iron Metabolism: From Mechanism to Therapy Potential; Trends Mol Med 2016, 22, 1077-1090. http://dx.doi.org/10.1016/j.molmed.2016.10.005
[8] Abergal, R., Clifton, M., Pizarro, J., Warner, J., Shuh, D., Strong, R., and Raymond, K. The Siderocalin/Enterobactin Interaction: A Link between Mammalian Immunity and Bacterial Iron Transport. JACS, 2008, 11524-11534.
[9] “Anthrax Stealth Siderophores.” Structural Biology Knowledgebase, NIH. http://doi:10.3942/psi_sgkb/fm_2012_6
