EXECUTIVE SUMMARY: Metal chaperones (or Metallochaperones) are small proteins responsible for transporting metals–more specifically, copper(I) and zinc(II)–through the cellular environment. [1] Scientists originally believed that metals entered the cell to bind to proteins via passive diffusion. [1] In recent years, however, scientists have discovered that these metals are present at extremely low intracellular concentrations and that a transport/delivery vehicle is necessary to prevent their scavenging or binding by unintended molecules. [1] As of now, most studies of metallochaperones have focused on those of copper(I) and zinc(II) and have emphasized the importance of the CXXC motif and cysteines in binding. [1] While three different copper(I) metallochaperones have been investigated at some length, the potential existence and binding properties of zinc(II) metallochaperones are still largely uncharacterized. [1]
Contents
click me to jump down a line!!
Copper(I) Metallochaperones
Three families of copper chaperones are currently known; Axt1, CCS, and Cox-family chaperones. All of these chaperones are thought to bind copper using two cysteine residues, typically through a binding motif with sequence CXXC (C = cysteine, X = any residue). [1] Atx1 or Atox1 (in yeast and humans, respectively) transports copper to vesicles in the secretory pathway, eventually contributing to metallation of membrane proteins. [1] CCS transports copper molecules to SOD1, an enzyme that utilizes copper in destroying superoxide radicals. [1] Finally, the Cox-family of chaperones are responsible for the delivery of Cu(I) to cytochrome c oxidase, the final enzyme in the mitochondrial respiratory chain. [1]
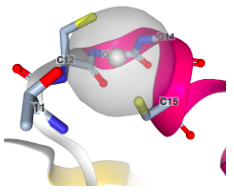
X-ray crystallographic structure of Atx1 copper binding site shown with a silver ion (proteins are sometimes easier to crystalize with different ions bound). Silver is bound in a linear fashion by two cysteine residues spaced by two amino acids (W. Wei, F. Wang, J. Zhao (2017), Structure of tetrasilver bound to human copper homeostatic proteins atox1 at 1.7 Angstroms resolution Prepublication).
Copper(I) in the Cell
In the cell, copper is involved many processes, including oxidative phosphorylation and regulation of oxidative stress. [2] Copper’s biological roles are influenced by its potential oxidation states and its cellular concentration. Copper can easily shuttle between its (I) and (II) oxidation states, making it both a useful cofactor in redox reactions and a highly toxic substance, able to generate harmful radical oxygen species through a process known as the fenton reaction. [4]
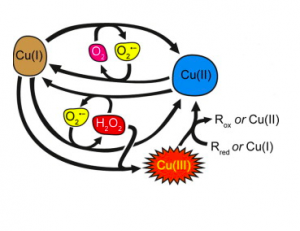
The Fenton Reaction. In this process, Cu(I) is reduced to Cu(II) and Cu(II) is oxidized to Cu(I). The radical oxygen species produced is superoxide. (Pham, et al.) [16]
In order to mitigate this cytotoxicity, it is important that cells keep free copper ions to attomolar concentrations – less than one atom per cell. [5] The result of this scarcity is that even proteins with incredibly high binding affinity for Cu(I) need an efficient source for their metal cofactor. Copper metallochaperones act as this source, aiding in the transport of Cu(I) through the cellular medium and protecting the ion from sequestration by unintended molecules. Such processes are incredibly important, as deficiencies in copper transport and regulation can lead to neurodegeneration and other diseases. [6]
Copper(I) Peptide Models
As it is often difficult to determine the binding interaction between a full protein and a bound metal ion, examining synthetic peptide models of partial Cu(I) metallochaperones can provide insights into how metallochaperones might bind Cu(I) in vivo.
For a study in 2013, Cu(I) was reacted with two cyclic peptides under acidic (pH~3.0) and basic conditions (pH~8.5). [4] Although both contained the CXXC motif, one peptide had a threonine, while the other had an aspartate. [4] The threonine was exchanged for an aspartate because the latter amino acid is found in the binding motif of a metallochaperone (ZntA) that binds to Zn(II). [4] Under acidic conditions, Cu(I) bound to aspartate and cysteine. [4] The binding was different under basic conditions and therefore pH dependent, but the study did not provide any further details. [4]
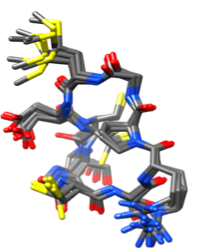
This is the Cu(I) peptide under acidic conditions. The structure is made up of 10 superimposed structures based on NMR data. (Shoshan, et al.)
Cu(I)’s bond to aspartate is noticeable because aspartate is present in the binding motif of Zn(II) metallochaperones but not in Cu(I) metallochaperones. [4] There are several lessons to be learned from this unusual result. First, in vivo, this binding might inhibit the metallochaperone’s release of Cu(I), so the cell may have mechanisms to prevent Cu(I) delivery to aspartate-containing metal binding proteins. [4] Second, it shows the importance of amino acid neighborhoods in metallochaperone binding. Although the CXXC motif remained constant throughout the two binding motifs tested, Cu(I) had different binding depending on the amino acids outside of this motif. Third, the fact that Cu(I) bound to aspartate in a metallochaperone-like fashion indicates that binding to aspartate could also be a reasonable strategy for Zn(II). [4]
Atx1
Atx1 is the metallochaperone responsible for the delivery of Cu(I) to Ccc2a. Ccc2 is a metal transport protein that sits on the surface of vesicles in the secretory pathway and drives copper into vesicles using the hydrolysis of ATP. [1] Once in the vesicle, copper can metallate proteins that are destined for cell membranes. This process aids in the metallation of deep, solvent-inaccessible pockets.
The Atx1 contains the conserved CXXC copper binding domain. Its CXXC motif is located at the beginning of an α-helix, making it more accessible for iron binding and donation. Atx1 is approximately 7 kDa and has a pyramid-like βαββαβ (β = beta sheet, α = alpha helix) tertiary structure with the copper metal at its “vertex.” Atx1 selectively binds Ccc2 by forming a dimer at the structurally similar Cu binding domain of Ccc2. A solved solution structure indicates that binding of this heterodimer is mediated by additional proteins called Atx1-Cys15 and Ccc2a-Cys13. Complementary surface electronics of the Ccc2 and Atx1 proteins, mainly comprising ionic residues, likely aids in selectivity of the interaction. [7]. Copper transfer occurs slowly, due largely to the similar binding pockets of the two involved proteins. A Ccc2 cysteine thiolate attacks the CuAtx1 coordination complex, forming a three-coordinate copper. One thiol from the Atx1 sheds the Cu-bond. Then the second Ccc2 cysteine completes the formation of the CuCcc2 system by attacking the copper ion, kicking off the Atx1 cystine [7].
It is important to note the nearly identical coordination motifs of Ccc2 and Atx1. Upon formation of the CuCcc2 coordination system, the system is nearly identical to the starting state. This is reflected in a equilibrium constant of ~1.5, suggesting that Cu Atx1’s role in the cellular environment may be less the transport of ions and more the protection of a scarce resource from a hyper coordinating system. Models of the binding sites, however, imply that there is a small increase in free energy upon copper transfer because of the change from a disordered to ordered binding site [8].
CCS
CCS, the copper chaperone for SOD1 (superoxide dismutase), is required to insert Cu(I) into SOD1. [9] SOD1 is a protein responsible for scavenging superoxide anion radicals in the cell through a reaction involving copper redox. [9] Interestingly, SOD1 is also requires a Zn(II) in order to be activated. [1] CCS delivers approximately 15% of available Cu(I) in the cell to saturate SOD1. [9] At 30-32 kDa, CCS is currently the largest known metallochaperone (Atx1 and the Cox-family are both about 7-8 kDa). [1] However, CCS’s size still pales in comparison to transferrin’s 80 kDa. [10]
CCS is unique because of its three functionally distinct domains. [1] Similar to Atx1, Domain I contains a CXXC domain. [1] Due to the presence of the CXXC domain, it was originally assumed that Domain I was essential to CCS activity. [1] Further studies showed that Domain I is not essential (it is, in fact, absent in Drosophila) but is used to maximize CCS activity when limited Cu(I) is available [6]. Unlike Domain I, Domains II and III are essential to CCS activity. [1] Domain II physically interacts with SOD1 to transfer Cu(I) to the protein, and Domain III, with a CXC binding motif (note the similarity to CXXC), is responsible for inserting Cu(I) into SOD1’s active site. [1] Notably, the three domains of CCS and SOD1 have a similar structure, further indicating the importance of partner recognition in transferring Cu(I) from a metallochaperone to a protein. [1]
Although exact mechanism of Cu(I)’s transfer from CCS to SOD1 is not yet completely understood, one has been proposed. [11] It is likely that the transfer includes the formation of a disulfide bond between CCS and SOD1, similar to the disulfide bond formed during the transfer of Cu(I) from Atx1 to Ccc2. [11] It is also thought that an oxidation of the nearby cysteine residues of CCS might be necessary to help drive the reaction forward. [11] The reaction requires this extra drive because it is possibly an unfavorable transfer for Cu(I): when coordinated to CCS, Cu(I) is bonded to thiolates, but when it is transferred to SOD1, it enters a histidine environment. As Cu(I) is a soft metal and the sulfur atom of cysteine is softer than the nitrogen atom of histidine’s imidazole, this is not a reaction Cu(I) could undergo passively. [11] Cu(I) transfer and CCS unbinding occurs before the addition of Zn(II) to SOD1, indicating that CCS may bind to a partially folded SOD1 protein; indeed, it is hypothesized that the binding of these two proteins is mediated by the mobile loop elements of a partially folded SOD1. Disulfide bond formation between the two proteins is not seen when CCS-Cu(I) is incubated with SOD1-Zn(II) [6].
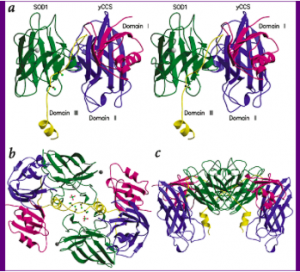
CCS binding to SOD1, from four different vantage points. Note the three binding motifs of CCS and the structural similarities between CCS and its target protein (Lamb, et al.).
Cox-Family
Cox-proteins, including Cox17, are involved in the delivery of copper to cytochrome c oxidase (CcO) at two Cu-redox centers (conflicting characterizations of the ligand-metal ratio have been published with values up to 4:1 Cu to protein [cit]). CcO is the final protein in the electron transport chain, and it is fundamentally important for the metabolic health of all eukaryotic organisms [2]. Yeast knockouts of Cox17 show deficient respiration and an inability to grow on oxidative carbon sources, and Cox17 knockouts in mice are lethal [12, 13]. CcO binds three copper ions: two near the surface of the mitochondrial inner membrane and one deep in the hydrophobic core. Because of this, two distinct proteins are needed to donate to the apo (non-metallated) form of the protein. The schematic representation below shows the pathway copper uses as it moves through the mitochondria to CcO.
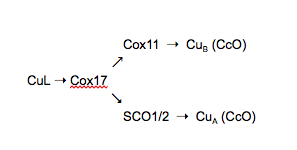
CuL, a small peptide bound to Cu, is the first identified member of this metallation pathway. It is unknown where in the cell CuL obtains its Cu or whether it aids in the transport Cu into the mitochondria. CuL donates one copper (I) ion to Cox17. In mammalian cells, Cox17 transfers Cu(I) to three downstream proteins: Cox11 and Sco1 and 2. Sco1/2 are localized in the inner membrane of the mitochondria, with copper-binding domains protruding into the intermembrane space. In vitro studies have shown direct transfer of Cu(I) from Cox17 to its downstream targets, which then transfer Cu(I) to CcO. The purported binding between Cox17 and these targets is the likely mechanism by which copper is selectively delivered to CcO [6].
Human Cox17 is a protein of 63 residues and has a twin CX(9)C binding domain [6]. Characterization of Cox17’s binding of Cu has been difficult due to the inability to isolate the metallated protein. It has been found, however, that there are 6 highly conserved cysteine residues within the Cox17 sequence that may be important for stabilizing its structure. Isolation of the protein shows 3 disulfide bonds, and treatment of the protein with reductants that simulate the environment of the mitochondrion gives two products: a peptide with 2 S–S bridges and the fully reduced apoprotein. [6]
Zinc(II) Metallochaperones
As of now, no Zn(II) metallochaperones are known. [14] However, due to the similarities between Cu(I) and Zn(II), it is likely that Zn(II) metallochaperones exist like Cu(I) metallochaperones. [14] In order to examine what potential Zn(II) metallochaperones might look like, it is possible to consider what is known about Zn(II) homeostasis and transport in the cell and various models of potential Zn(II) binding to metallochaperones. [14]
Zinc(II) Homeostasis
Zn(II) homeostasis in cells involves the uptake and efflux of Zn(II)–in other words, its transport. [14] One protein that is involved in this cycle is YiiP, a protein that transports transition metal ions. [14] Yiip is a dimeric protein: its subunits are separated, but its carboxy terminus tails are associated. [14] Each subunit binds four Zn(II) ions, but it is likely that Yiip can bind to excess Zn(II) for stabilization and activation. [14] Such a method of binding to additional Zn(II) is a method of controlling Zn(II) concentrations in the cell through activity rather than through gene expression. [14] This not only allows for finer control; it also accounts for the fact that there is often an excess of Zn(II) in the cytoplasm due to the non-specificity of metal transporters into the cell. [14] Although it is not yet completely clear how YiiP and Zn(II) interact in vivo, YiiP’s metallochaperone-like structure and function indicate that studying YiiP and similar proteins at greater length will yield more information regarding the potential for Zn(II) metallochaperones. [14]
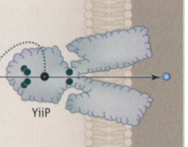
The YiiP protein. The green dots indicate where Zn(II) can bind to the protein (Nies).
Zinc(II) Peptide Models
Investigating how Zn(II) binds to peptide models of metallochaperones provides further insight into how Zn(II) might bind to potential metallochaperones in vivo. In a study conducted by Shoshan, et al. in 2013, Zn(II) was reacted with cyclic peptides with an aspartate mutant strain of the Cu(I) wild-type metallochaperone binding motif. [4] The sequence of this mutant strain was MDCSGCSRPG. It is important to note that in the mutation, the integrity of the CXXC motif common to Cu(I) metallochaperones was preserved. [4]
Zn(II) had different binding under acidic and basic conditions. [4] Under acidic conditions, Zn(II) bound to a methionine and two cysteines, while under basic conditions, it bound to one methionine and one cysteine, resulting in a lower coordination number. [4] It is likely that under basic conditions, Zn(II) also bound to a deprotonated H2O ligand. [4] The amino acids Zn(II) was likely bound to was again determined by measuring the distances between the atoms available for binding in the amino acids. [4] These atoms were deemed reasonable for binding to Zn(II) if they less than 8Å apart. [4]
The most interesting result of this study was the fact that Zn(II) did not bind to aspartate, even though aspartate was inserted into the binding motif specifically because Zn(II) is known to bind to aspartates in transporters in vivo. [4] Even more, the fact that Cu(I) bound to aspartate when reacted with this mutant strain indicates that it was a reasonable–and perhaps even favorable–amino acid to bind to. [4] Zn(II)’s lack of binding to aspartate is only rendered more surprising given hard/soft acid/base theory. [4] As Zn(II) is a harder metal, it would be expected that it would prefer to bind to the harder CO2– group of aspartate rather than the softer S– of methionine. [4] Yet, perhaps Zn(II) strongly prefers coordination numbers above two–more than it prefers binding to a harder group. [4] This type of coordination requires cyclizations and severe steric strain, which might only be possible near the sulfur of methionine. [4] Nonetheless, the fact that Zn(II) bound to these peptides in a similar manner to Cu(I) indicates that it could bind to a Cu(I)-like metallochaperone. [4]
Potential Binding Structures
In 2013, Badarau, et al. used X-ray crystallography to investigate structural mechanisms of Zn(II) metallochaperones. [15] The authors constructed chimeras of copper- and zinc-binding proteins to measure metal-specific binding affinities and obtain crystal structures. [15] The addition of the N terminus of ZiaA (a Zn(II) exporter) to two different copper chapperones (Atx1 and Ccc2) to make Atx1ZiaA(N) and Ccc2ZiaA(N). [15] Though the CXXC pattern was conserved between all residues, the non-cysteine residues differed (see figures below), pointing to possible structural mechanisms of selectivity [15].
Metal ions were incubated with Atx1, Atx1ZiaA(N), Ccc2, and Ccc2ZiaA(N). [15] The chimeric proteins showed significant increases in Zn(II) binding affinities but only slight increases in Cu(I) affinity. [15] Atx1-Cu(I) binding affinity was 4.7×1017 and increased marginally to 5.6×1017 upon addition of the ZiaA N terminus, while Zn(II) binding affinity increased from 7.2×108 to 2.5×1010. [15] So far, crystal structures of wildtype ZiaA have been elusive. [15] Chimeric proteins, however, yielded a putative tetrahedral binding structure, with Zn(II) bound to two cysteines of each dimerized Atx1ZiaA(N). [15] Zn(II)-S distances of ~2.3-2.4Å and bond angles of 101-121˚ agree with a tetrahedral structure [15].
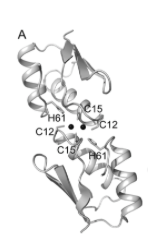
Cu(I) bound to Atx1 with the binding motif IACEAC (Badarau, et al.)

Zn(II) bound to Atx1ZiaA(N) with the binding motif MDCTSC (Badarau, et al.)
Similarly to Atx1, Ccc2-Cu(I) binding affinity increased slightly from 7.8×1016 to 8.5×1016 upon addition of the ZiaA N terminus, while Zn(II) affinity increased drastically from 4.2×107 to 1.7×109. [15] Two crystal structures were obtained under different crystallization conditions, indicating that there may be some flexibility or dynamism in binding. [15] One crystal structure indicated the coordination of Zn(II) by two cisteins, an aspartate, and a histidine, with bond distances of ~2.3Å (Zn(II)-S), 1.95Å (Zn(II)-O), and 2.05Å (Zn(II)-N). [15] Bond angles of 104-120˚ agreed with a tetrahedral structure. [15] The second crystal showed replacement of the aspartate with a water ligand, maintaining the tetrahedral structure with a Zn(II)-O bond distance of ~2.1Å. [15] These structures together indicated a possibly dynamic role of aspartate in Zn(II) binding and release, as well as a possible unique mechanism for Zn(II) selectivity. [15]
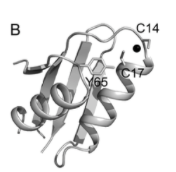
Cu(I) bound to Ccc2 with the binding motif MRCAAC (Badarau, et al.)

A. Zn(II) bound to two cysteines, an aspartate, and a histidine with the binding motif MDCTSC.
B. Zn(II) bound to two cysteines, a histidine, and a water ligand with the same binding motif as A. (Badarau, et al.)
These chimeras showed the influence of non-cysteine residues on Zn(II) binding affinity. [15] It is important to note, however, that all wild-type and chimeric proteins had a higher affinity for Cu(I) than Zn(II), suggesting other, non-structural mechanisms of ion selectivity. [15]
See More
Here are three videos that describe various aspects of metallochaperones that we have touched on in greater detail!
1. This video details Atx1 activity in the cell. It provides an extremely comprehensive view of Atx1, moving beyond Atx1’s functions to go into hard/soft acid/base theory, electron count, ligand field stabilization energy, and kinetic lability.
2. 29:00-34:00. This video introduces metallochaperones in the context of cell compartments and transport through them. It outlines the roles of Atx1, including its localizations, and how copper is transferred without being released as a free ion. It also dives into Wilson’s disease and mechanisms of drug treatment. Diagrams and biological references would give students a context for why Atx1 is important, as well as introduce them to themes of compartmentalization and localization that are critical for metallochaperone function.
3. This video explains the biological importance of SOD1, outlines metal’s role in electron transfer, details redox reactions, and investigates SOD1 protein structure in relation to metal binding. This video will be useful for students because of its simplified diagrams, accessible discussion of chemical topics like redox reactions and protein biochemistry.
References
- O’Halloran, T.V.; Culotta, V. “Metallochaperones.” In Biological Inorganic Chemistry; Bertini, I.; Gray, H.B.; Stiefel, E.; Valentine, J.S., Ed. University Science Books, 2006; pp. 166-174.
- J. Robinson, D. R. Winge (2010), Copper Metallochaperones, Annu. Rev. Biochem. 79, 537–562.
- Ridge, P.G., et al. 2008. Comparative genomic analyses of copper transporters and cuproproteins reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS ONE 3:e1378.
- Shoshan, M.S.; Shalev, D.E.; Tshuva, E.Y. Peptide Models of Cu(I) and Zn(II) Metallochaperones: The Effect of pH on Coordination and Mechanistic Implications. Inorganic Chemistry. [Online] 2013, 52, 2993-3000 doi: dx.doi.org/10.1021/ic302404w (accessed February 15, 2017).
- Finney, L.A.; O’Halloran, T.V. Transition Metal Speciation in the Cell: Insights from the Chemistry of Metal Ion Receptors. Science. [Online] 2003, 300, 931-936 http://www.jstor.org/stable/3833920 (accessed February 15, 2017).
- Robinson, N.J. and Winge, D.R. 2010. Copper Metallochaperones. Annu Rev Biochem 79:537-62.
- Banci, L., et al. 2006. The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction. Nat Chem Biol 2:367-68.
- Huffman, D.L. and O’Halloran, T.V. 2000. Energetics of copper trafficking between the Atx1 metallochaperone and the intracellular copper transporter Ccc2. J Biol Chem 275:18611-14.
- Rae, T.D.; Schmidt, P.J.; Pufahl, R.A.; Culotta, V.C.; O’Halloran, T.V. Undetectable Intracellular Free Copper: The Requirement of a Copper Chaperone for Superoxide Dimutase. Science. [Online] 1999, 284, 805-808 http://www.jstor.org/stable/2898319 (accessed February 15, 2017).
- Chung, M. Ching-Ming. Structure and Function of Transferrin. Biochemical Education. [Online] 1984, 12, 146-154, http://onlinelibrary.wiley.com/store/10.1016/0307-4412(84)90118-3/asset/5690120402_ftp.pdf?v=1&t=j13woc13&s=6ca9d581aed7369f13571bc56bb762ee42569dc4.
- Lamb, L. A.; Torres, A.S.; O’Halloran, T.V.; Rosenzweig, A.C. Heterodimeric Structure of superoxide dismutase in complex with its metallochaperone. Nature Structural Biology. [Online] 2001, 8, 751-755, doi: 10.1038/nsb0901-751 (accessed February 28, 2017).
- Glerum, D.M., et al. 1996. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 271: 14504-9.
- Takahashi, Y., et al. 2002. Mammalian copper chaperone Cox17 has an essential role in activation of cytochrome c oxidase and embryonic development. Mol. Cell. Biol. 22: 7614-21.
- Nies, D.H. How Cells Control Zinc Homeostasis. Science. [Online] 2007, 317, 1695-1696, http://www.jstor.org/stable/20048413 (accessed February 24, 2017).
- Badarau, A., et al. (2013). Investigating the role of zinc and copper binding motifs of trafficking sites in the cyanobacterium Synechocystis PCC 6803. Biochemistry 52:6816-6823.
- Pham, N.A.; Xing, G.; Miller, C.J.; Waite, T.D. Fenton-like copper redox chemistry revisited: Hydrogen peroxide and superoxide mediation of copper-catalyzed oxidant production. Journal of Catalysis. [Online] 2013, 54-64, doi: 10.1016/j.jcat.2013.01.025.
