Sulfite Oxidase
Molybdenum (Mo) sulfite oxidase is an enzyme primarily responsible for catalyzing the conversion of sulfite to sulfate, as excess sulfite in the body can result in various forms of neurodegeneration [1]. With two metal centers, Mo sulfite oxidase undergoes redox reactions involving its molybdenum center and the iron from its heme group. Although several synthetic models of the enzyme have been created, there is yet to be one that successfully replicates both its structure and its function. This points toward a potential future goal in better understanding Mo sulfite oxidase.
- Mo’s Role in Biology
- Structure
- Function
- Synthetic Models
Mo’s Role in Biology
Molybdenum is the only second-row trace metal that acts as a cofactor in human cells [1]. Most enzymes that utilize molybdenum do so by binding a molybdenum cofactor (MoCo, Figure 1) synthesized from a cofactor molecule called pterin. Molybdenum enzymes are critical for global nutrient cycles, including those of nitrogen and nitrogen [1]. They act by transferring an oxygen atom, rather than an oxygen ion, to or from their substrates.
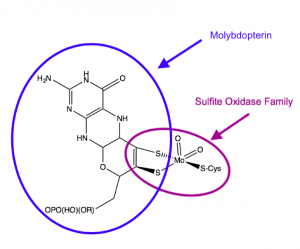
Figure 1. Structure of the molybdenum cofactor utilized by sulfite oxidase. From Schwarz, 2016.
Humans have four enzymes that utilize MoCo: sulfite oxidase (SO), xanthine oxidoreductase, aldehyde oxidase, and mitochondrial amidoxime reducing component [2]. Genetic studies have shown that SO is the most important of these enzymes, leading to disease phenotypes like neurodegeneration and seizures if mutated [2]. SO is an inner mitochondrial membrane protein involved in the breakdown of sulfur-containing amino acids like methionine and cysteine [1]. When methionine and cysteine are catabolised, they form multiple intermediate molecules, including sulfite. Sulfite (SO32-) enters the mitochondrial intermembrane space and encounters SO. SO then transfers an oxygen atom from water to convert sulfite into sulfate (SO42-) (Figure 2).
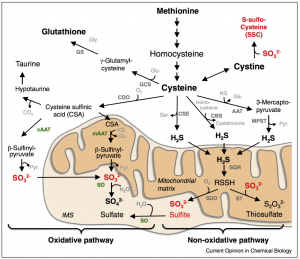
Figure 2. Breakdown of sulfur-containing amino acids. Sulfite oxidase (SO, labeled in green) is present at the inner mitochondrial membrane and converts sulfite to sulfate as part of the oxidative pathway shown in the lower left.
A mutation in SO leads to a buildup of sulfite, which can cause damage to the cell by producing reactive oxygen species (ROS) and causing DNA or membrane damage. Sulfite is also dangerous in that sulfite and its related metabolites can interfere with glutamate-related processes: sulfite can inhibit glutamate dehydrogenase, an important source of cellular ATP, and sulfite-derived S-sulfocysteine (SSC), formed in the blood, can activate MDMA receptors by binding the glutamate site [1]. This activation of NDMA is thought to be the cause of seizures and neurodegeneration in patients with SO mutations [1].
Structure
In examining the structure of Mo sulfite oxidase, its two metal centers, two oxo groups, and flexibility are key. Mo sulfite oxidase is a α2 dimer that weights about 106 kDa [3]. Each subunit has a b-type cytochrome heme group and an Mo center that are connected by a 10-residue loop [3, 4]. The loop is flexible, weak, and relatively easily cleaved by enzymes [3]. Interestingly, the iron centers of the heme groups and the Mo centers are farther than would be expected, with a 32 Å distance between them [3]. Usually, atoms need to be between 20-25 Å to interact. Despite this distance, the rate of reaction between Mo and iron is relatively fast, at ~103 s-1 [3]. Given the 32 Å between them, the expected rate of reaction would be <100 s-1, which would be significantly slower [3]. The fact that Mo and iron can react so quickly despite their distance indicates the potential importance of the flexible 10-residue loop–perhaps this loop can change conformation to allow Mo and iron to interact so efficiently [3].
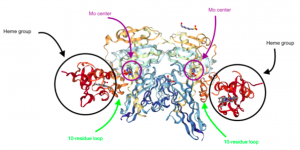
Figure 3. The structure of Mo sulfite oxidase. Note the heme groups, the Mo centers, and the 10-residue loops connecting the two metal centers. (Adapted from Protein Data Bank. 1SOX: Sulfite Oxidase from Chicken Liver. http://www.rcsb.org/pdb/ngl/ngl.do?pdbid=1SOX&bionumber=1 (accessed April 21, 2017)).
The Mo center is also important to examine, as its oxygen atoms are involved in reactions as well. The MoO2 center has square pyramidal geometry, with the O=Mo=O angle at approximately 105˚ [3]. One of the Mo=O groups is axial, while the other is equatorial, and the Mo=O bond distance is about 2.3 Å [3]. The electronics of the O=Mo=O bond indicate the bond’s strength. Mo and O form 5 bonding orbitals and one non-bonding orbitals, resulting in a bond order of 2.5 [3]. The π bonding orbitals are formed by constructive interference between the lobes of Mo’s 4d orbitals and O’s p orbitals, and the π* orbitals are formed through destructive interference of the same lobes (Figure 4).
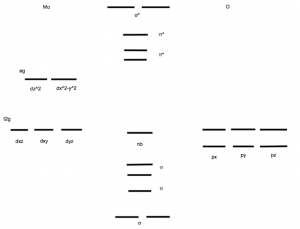
Figure 4. A molecular orbital diagram of the Mo=O bonds. Both oxygen atoms have full p orbitals because they are oxo species. Mo has no 4d electrons due to its positive charge. Mo=O forms two sigma orbitals, three π orbitals, and one nonbonding orbital. (Adapted from McMaster, J.; Garner, C..D.; Stiefel, E.I. Molybdenum Enzymes. In Biological Inorganic Chemistry; Bertini, I.; Gray, H.B.; Stiefel, E.I.; Valentine, J.S. University Science Books, 2007, p. 525.)
In addition to the oxo groups, Mo is bonded to its cofactor, molybdopterin (MPT) through a dithiolene ligand. The dithiolene ligand is especially stable due to its C=C backbone, delocalized π system, ands ability to donate π electrons to the Mo center [3]. The MPT ligand itself also has several redox centers, mainly composed of sulfurs and nitrogens which allow it to undergo reactions separate from the main reactions occurring at the Mo center [3]. The dithiolene ligand is also important in that its resonance structures may aid the molybdenum center in stabilizing electron density (Figure 5).

Figure 5. The structure of the Mo center in Mo sulfite oxidase.
Function
The function of Mo sulfite oxidase is to catalyze the conversion of sulfite to sulfate, which occurs through several intramolecular reactions [3]. The overall equation (Eq. 1) for this reaction indicates the change from sulfite to sulfate and the fact that the iron from the heme group (and therefore the Mo as well) undergoes a redox reaction [3].
SO32- + H2O + 2Fe(III)Cyt c → SO42- + 2H+ + 2Fe(II) Cyt c (Eq. 1)
The reaction cycle essentially has two main pieces: an oxygen atom transfer and redox reactions between Mo and Fe [3]. The reaction cycle begins by the sulfur of sulfite acting as a nucleophile and abstracting an oxygen atom (not an ion) from the Mo(VI) center [3]. As an H2O group attacks the Mo center, the sulfate is released [3]. Next, Fe(III) oxidizes Mo(IV) to become Mo(V) as a proton leaves the H2O group [3]. In the meantime, Fe(II) is oxidized, becoming Fe(III), which allows it to oxidize Mo(V) to become Mo(VI) [3]. At the same time, a proton is released, allowing the reaction cycle to continue (Figure 6) [3].
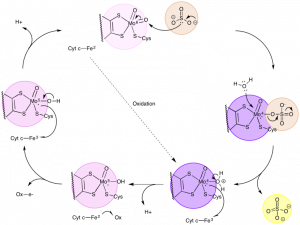
Figure 6. The catalytic cycle, which includes the conversion of sulfite to sulfate and the oxidation/reduction of Mo and Fe. (Adapted from McMaster, J.; Garner, C..D.; Stiefel, E.I. Molybdenum Enzymes. In Biological Inorganic Chemistry; Bertini, I.; Gray, H.B.; Stiefel, E.I.; Valentine, J.S. University Science Books, 2007, p. 537.)
Synthetic Models
Given SO’s unique efficient atom transfer capacity, researchers have been looking to develop catalysts capable of similar reactions in a multitude of solvent conditions with similar turnover and efficiency.
The first systems which were capable of replicating molybdenum mediated oxygen transfer fell victim to Mo(IV) and Mo(VI)’s propensity to form a stable μ-oxo Mo(V) dimers in solution when unprotected by the enzymatic chaperone—a significant barrier to the development of an Mo catalyst system. This problem was overcome by the use of bulky ligand structure to prevent association between Mo species in solution in the Berg-Holm model. (fig. 7) [5]
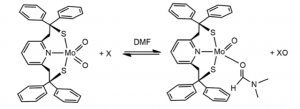
Figure 8. The Burg-Holm catalyst system.
While the Berg-Holm catalyst performed OAT, the ligand structure and oxygen regeneration step of the reaction are significantly different than the model enzyme.
The use of a dithiolene ligand in SO-mimetic catalyst is difficult to achieve in a way that mimics the structure of the SO’s reactive core, as the preferred coordination sphere of Mo(VI) is a hexaco-ordinate system, and the two terminal oxo species leave four binding sites which are occupied easily by two dithiolene ligands. The reaction mechanism in figure 9 was used to achieve a model of the SO core, with reasonable agreement in the bond angles and lengths between the model system and the enzyme. [5]
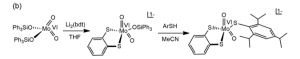
Figure 9. The synthesis a structural analogue to SO
Regrettably, the structure in figure 9 did not demonstrate reactivity on sulfite or sulfite analogues like PPh3.
Success in a functionally and structurally analogous synthetic model came with a bis- dithiolene ligand structure (fig. 10). Further, this model was capable of regenerating itself from H2O in solution by transferring electrons to K2[Fe(II)(CN)6]. Importantly, the issues in developing a model catalyst system for this enzyme highlight the tradeoffs that often need to be made between structurally and functionally accurate models. [5]
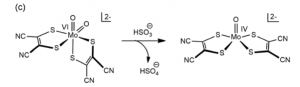
Figure 10. Functional model that exhibits a dithiolene ligand structure and reactivity.
References
[1] Schwarz, G. (2016). Molybdenum cofactor and human disease. Curr Opin Chem Biol 31:179-87.
[2] Reiss, J. (2016). Molybdenum cofactor and sulfite oxidase deficiency. Metabolomics 6(3):184.
[3] McMaster, J.; Garner, C..D.; Stiefel, E.I. Molybdenum Enzymes. In Biological Inorganic Chemistry; Bertini, I.; Gray, H.B.; Stiefel, E.I.; Valentine, J.S. University Science Books, 2007.
[4] Hille, Russ. Structure and Function of Mononuclear Molybdenum Enzymes. JBIC. 1996, 1, 397-404.
[5] Majumdar, A; Structural and functional models in molybdenum and tungsten bioinorganic chemistry: description of selected model complexes, present scenario and possible future scopes Dalton Trans., 2014, 43, 8990
