Summary
[NiFe] hydrogenases are bacterial enzymes responsible for the heterolytic cleavage of hydrogen. The protein structure contains a NiFe active site which binds hydride, and three transport pathways for H,2, H2O, and electrons. H2 is split as part of a catalytic cycle that has been determined from synthetic model studies. [NiFe] hydrogenases could potentially be harnessed as an H2 synthesizer as an environmentally clean energy energy source.
Table of Contents
Summary
Context
Structure and Function
Active site and Catalytic Cycle
Proposed Canopy Model
Oxygen Sensitivity
References
Context
The burning of fossil fuels releases carbon dioxide and other greenhouse gases into the atmosphere, contributing to global warming. On April 21, 2017, the atmospheric CO2 concentration reached 410 ppm, a concentration so high that the earth has not reached this level in millions of years (Figure 1)(1,2). This rise in CO2 concentration correlates with the rise in temperature of the earth, resulting in various consequences, for example, the melting of the ice caps.
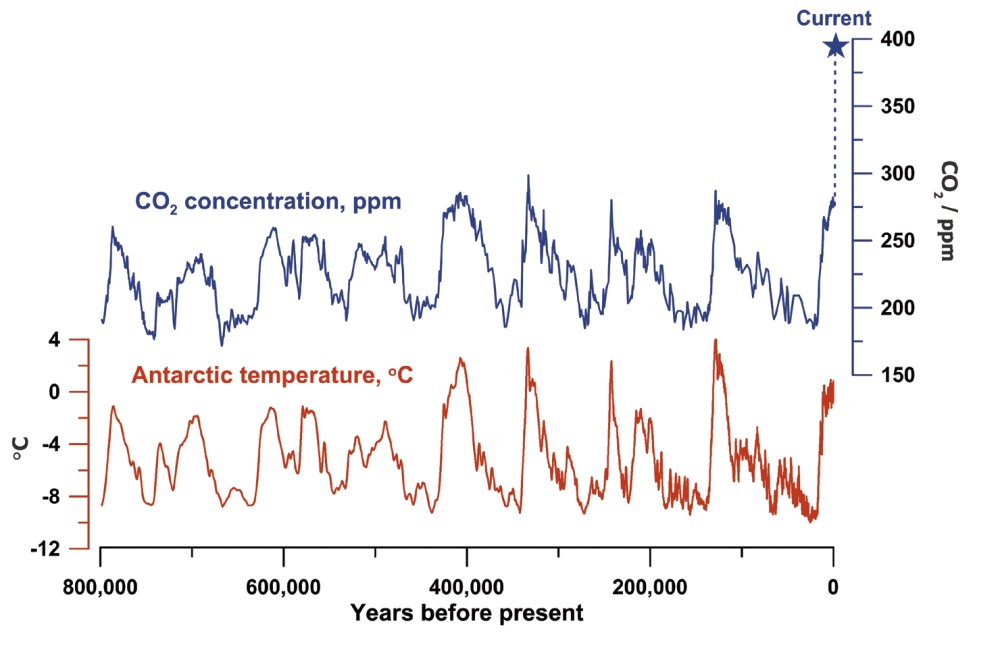
Figure 1: Historic atmospheric CO2 levels.2
Now more than ever, fossil fuels need to be abandoned and alternative energy sources need to be discovered. One promising alternative energy source is hydrogen gas. Hydrogen can be used in fuel cells to power motor vehicles. This fuel source is environmentally friendly because it yields only water as a byproduct. However, current means of producing H2 are unsatisfactory. Steam reforming, which produces H2 from methane (Scheme I) also produces CO2 as a byproduct.
Scheme I:CH4 + H2O⇌ CO +3H2
CO+H2O⇌ CO2+H2
Electrolysis of water is another alternative to producing H2, however this process is costly and difficult to scale up. Therefore, harnessing the power of biological catalysts to synthesize H2 is an attractive option. [NiFe] hydrogenases are a class of enzymes that catalyze the reversible reaction shown in Scheme 2:
Scheme 2: H2 ⇌ H– + H+ ⇌ 2H+ + 2e–
Therefore, the ability of [NiFe] hydrogenases to synthesize or break down H2 could be a useful tool for scientists developing alternative sources for H2.
Structure and Function
[NiFe] hydrogenases are enzymes that catalyzes the heterolytic splitting of H2 (or the reverse reaction) using a catalytic center that contains one nickel and one iron atom. Because these elements are relatively accessible to humans, the Ni-Fe catalytic center is of particular interest as a potential model for a biocatalyst.
[NiFe] hydrogenases are typically comprised of two subunits, a small subunit and a large subunit (some hydrogenases are membrane-bound, and contain a third subunit that is integral to the membrane). The small subunit contains three Fe-S clusters (two Fe4S4 and one Fe3S4) at close distances (about 10 angstroms apart). The large subunit contains the catalytic Ni-Fe center (Figure 2).
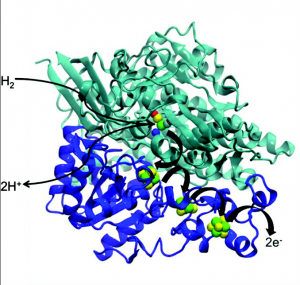
Figure 2: [NiFe] hydrogenase structure, showing large subunit in green and small subunit in blue. FeS clusters are shown in the small subunit in green and yellow (Sumner).
For the enzyme to split H2 the following must occur: H2 diffuses in toward the buried active site; it binds to the active site and is split; protons are channeled away from the active site toward the surface of the protein; electrons are channeled away from the active site, and ultimately to a redox partner (such as a cytochrome).
Three transfer pathways exist in the enzyme. The electrons flow away from the active site through the Fe-S clusters. The protons are channeled away via amino acid residue groups that can donate/accept protons and water molecules. The H2 diffuses in through gas tunnels, which are lined with hydrophobic amino acids (Figure 3). These gas channels were visualized in studies with krypton, a gas ideal for investigating gas tunnels because it is hydrophobic and large. Crystallizing the [NiFe] hydrogenases together with krypton and visualizing where the krypton was bound showed that there was a hydrophobic gas tunnel through which H2 can diffuse to the active site.
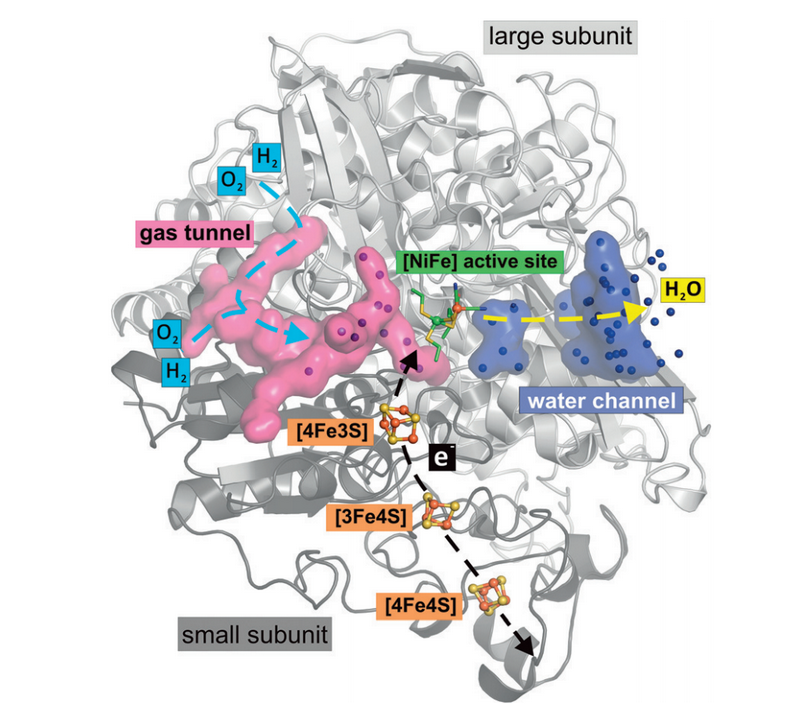
Figure 3: Hydrophobic gas tunnels, shown in pink, allow for gas diffusion to the active site (Kalms).
Active site and Catalytic Cycle
The Ni ion is coordinated by the thiolates of four cysteine residues (CX2C on the N-terminal side of peptide, CX2CX2H/R on C-terminal side), which are highly conserved. Two of the thiolates are bridging, also coordinating the Fe ion. The Fe ion is also coordinated by two CN– molecules and one CO molecule. These molecules are of biogenic origin (the CN– comes from carbamoyl phosphate). The cyanide and carbon monoxide molecules lock the Fe in a ferrous, low-spin state (Figure 4).
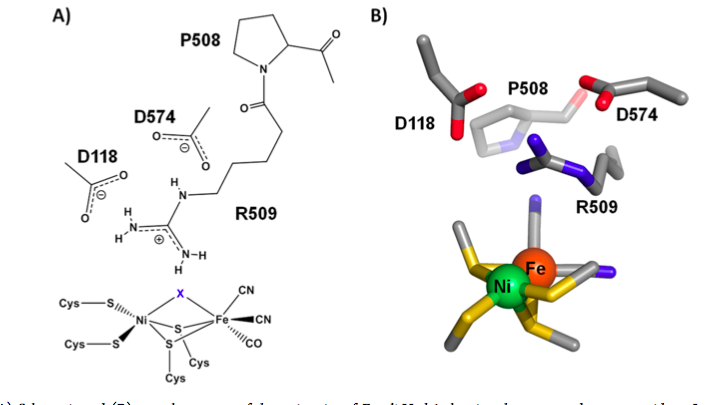
Figure 4. Active site of [NiFe] hydrogenase. R509 represents an arginine residue in the second sphere that potentially plays a role in catalysis. The atom labeled “X” represents either a briding hydride (during catalysis) or a bridging oxygen atom (in enzymes inactivated by oxygen) (Brooke).
The large variety of ligands (the thiolates are π-donors, the CO is a π-acceptor, and the CN– is a σ-donor) provides a flexible coordination environment, and allows for several stable redox states of the active site. This is important for the entry and exit of electrons at the active site.
During catalysis, there is also a bridging hydride ligand that is coordinated to both the Fe and the Ni atom. This is an important part of the catalytic cycle. The catalytic cycle, while not yet fully elucidated, involves three distinct redox states called Ni-R (fully reduced), Ni-SIa, and Ni-C. These sites can be separated through various spectroscopy techniques.
When the hydrogen molecule binds at the active site (Ni-SAa), it is split heterolytically, to a hydride and a proton. A basic group accepts the proton and the hydride becomes a bridging ligand to the Ni and Fe ions (forming the Ni-R state). There is debate whether the basic group is one of the cysteines in the inner sphere of coordination (this seems unlikely, however, because a thiol is not as effective a ligand). Then, the electrons are transferred from the hydride ligand to the Ni atom, and the resulting proton is released; the electrons are transferred away through the FeS clusters, reoxidizing the active site, and the original state (Ni-SIa) is reformed. In the video below, the hydride ligand is shown in a model as a white sphere between the nickel and iron atoms.
Proposed Canopy Model
There is compelling evidence that the basic group accepting the first proton is in the outer sphere of the active site. Mutating a strictly conserved arginine residue that dangles its guanidine nitrogen atom just above the Ni and Fe atoms (Figure 4) to lysine, which preserves the side chain length and basicity of the residue, results in a one hundred fold reduction of activity (Figure 5). This suggests that this residue plays a crucial role in catalysis, despite not being part of the inner coordination sphere of the active site.
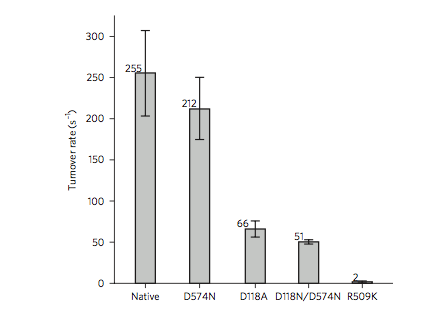
Figure 5. Mutating a strictly conserved arginine (R509) to lysine (K) results in a one hundred fold reduction of activity of [NiFe] hydrogenase (Evans).
One possible mechanism for the heterolytic splitting of hydrogen is that H2 is polarized by simultaneous binding to the Ni and Fe metals (lewis acids) and the guanidine N atom of the arginine residue (the base). This acts to polarize the H-H bond, such that one of the hydrogen atoms is left with both of the electrons (Figure 6).
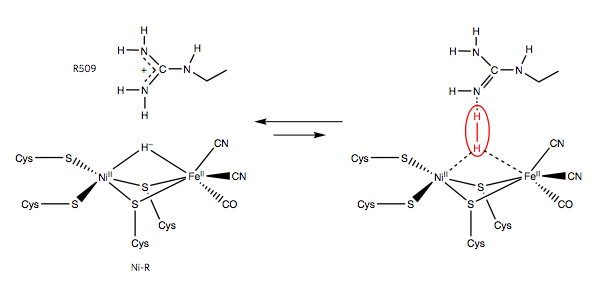
Figure 6. Proposed mechanism for hydrogen activation by the R509 arginine residue (Evans).
Oxygen Sensitivity
When designing a biocatalyst, it’s important to consider its stability in air! There are O2-sensitive and O2-tolerant subtypes of [NiFe] hydrogenases. The O2-sensitive subtype is inactivated in the presence of oxygen. A bridging oxygen atom permanently binds at the site where the hydride would bind during catalysis. The O2-tolerant subtype remains active in the presence of oxygen, and is therefore attractive as a model for biocatalysis. The O2 tolerance is due to the enzyme’s capacity to use use some of the electrons generated by H2 oxidation to reduce the O2 to water; four electrons are required for this process.
Studies suggest that O2 tolerance is related to the amount of gas tunnels that the hydrogenase contains; O2-tolerant enzymes have fewer gas tunnels than do O2-sensitive enzymes, which could lead to a lower rate of O2 diffusion to the active site. Additionally, O2-tolerant enzymes contain a special proximal [4Fe-3S]-6Cys cluster, which is in a superoxidized state, and can deliver two electrons to oxygen at once. This aids in the reduction of the bound oxygen to water and allows for reactivation of the enzyme.
References
- https://www.scientificamerican.com/article/we-just-breached-the-410-ppm-threshold-for-co2/
- https://royalsociety.org/~/media/Royal_Society_Content/policy/projects/climate-evidence-causes/fig3-large.jpg?la=en-GB
Brooke, E. J., et al. Biochemistry 2016.
De Lacey, A. L., et al. Chem. Rev. 2007, 107 (10), 4304-4330.
Evans, D. J.; Pickett, C. J., Chem. Soc. Rev. 2003, 32 (5), 268-275.
Evans, R. M., et al. Nat. Chem. Biol. 2016, 12 (1), 46-50.
Kalms, J., et al. Angew. Chem. Int. Ed. 2016.
Ogata, H., et al. J. Biochem. 2016, 160 (5), 251-258.
Figures
Figure 2: Reprinted with permission from Sumner, I.; Voth, G.A. Proton Transport Pathways in [NiFe]-Hydrogenase. The Journal of Physical Chemistry B 2012 116 (9), 2917-2926. Copyright 2012 American Chemical Society.
Figure 3: Reprinted with permission from Kalms, J., et al. Angew. Chem. Int. Ed. 2016.
Figure 4: Reprinted (adapted) with permission from Brooke, E.J.; Evans, R.M.; Islam, S.T.A.; Roberts, G.M.; Wehlin, S.A.M.; Carr, S.B.; Phillips, S.E.V.; Armstrong, F.A. Importance of the Active Site “Canopy” Residues in an O2-Tolerant [NiFe]-Hydrogenase. Biochemistry 2017 56 (1), 132-142. Copyright 2016 American Chemical Society.
Figures 5 and 6: Reprinted from Evans, R. M., et al. Nat. Chem. Biol. 2016, 12 (1), 46-50.
